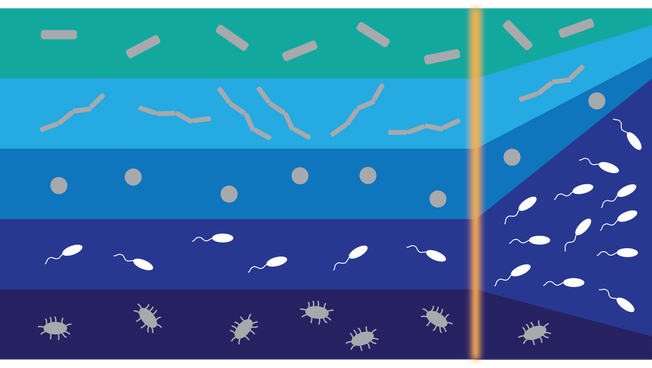
ASSESSING AND MANIPULATING CONSTRAINTS ON MICROBIAL NICHE BREADTH
Our lab focuses on the constraints and consequences of soil microbial niche breadth. Fundamentally, we aim to understand current and future microbial biogeography and the fitness and functional costs of different types of generalism. From an applied perspective, we address the most recalcitrant challenge of microbial management, which is the unpredictable in-field survival and function of beneficial microbes.
Soil microbes shape Earth’s biogeochemistry and primary productivity, but vary widely in their performance efficiency, meaning that even small shifts in microbial traits or composition can have large functional consequences across ecosystems. At the extreme, historical shifts have led to mass extinction events and reconstituted the atmosphere. As humans reconfigure global soils, including conversion of ~40% of arable land to agriculture, they alter the diversity and abundance of soil resources, stressors, and structures, and in turn, the eco-evolutionary landscapes that select for microbial traits and taxa. Agriculture has been hypothesized to shift microbial functions globally, through changes in microbial traits and by promoting microbes that thrive in low heterogeneity soils receiving high resource inputs. Although microbial lineages have diverged over billions of years, near-term environmental change can quickly reshape soil microbiomes through the rapid evolution of shallowly conserved traits, altered intermicrobial interactions, and species sorting.
Most of our current work addresses at least one of three key questions:
- How do diverse microbial lineages respond to new environments?
- How do complex microbial assemblages respond to new environments?
- How do interactions between microbial and environmental traits shape microbiome function?
We address our questions through active filtering of microbiomes, directed evolution, environmental manipulation, and observational studies. We extensively apply various omics techniques, which we combine with approaches from ecology, soil science, and microbiology.
Selected Past Projects
Specialist and generalist microorganisms in agricultural systems
Funded by the USDA Foundational Program (PI is TH Bell)
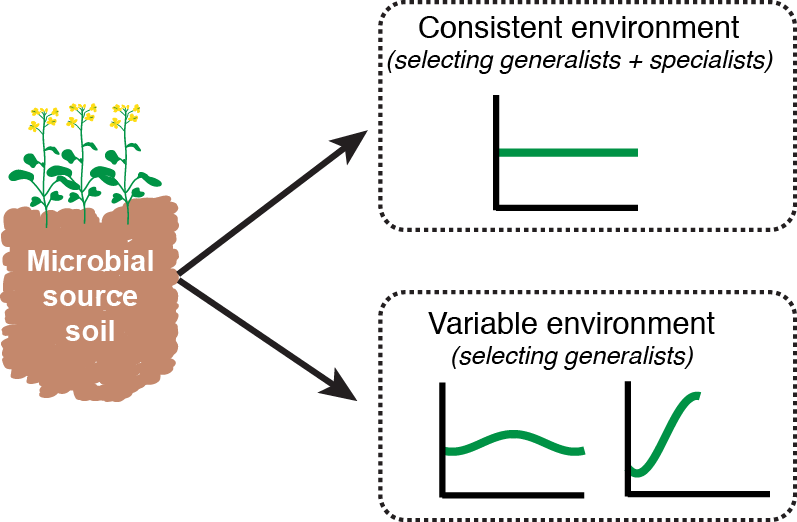 Agriculture typically reduces biodiversity relative to unmanaged environments, selecting against specialist organisms that survive under narrow conditions, while promoting generalists that tolerate a broader environmental range. Various management practices (e.g. nutrient applications) impose cyclical pressures that microorganisms must contend with, and that widen intra-annual variability in the environment. Shifting towards generalist-dominated communities could have landscape-scale consequences for key soil processes. In this project, we impose differing environmental gradients in custom in-soil selection systems, allowing us to collect generalists and specialists of different types. We then assess the taxonomy and function of these assemblages. Ultimately, we aim to augment functional and ecological annotations of sequence data, and better inform microbial management.
Agriculture typically reduces biodiversity relative to unmanaged environments, selecting against specialist organisms that survive under narrow conditions, while promoting generalists that tolerate a broader environmental range. Various management practices (e.g. nutrient applications) impose cyclical pressures that microorganisms must contend with, and that widen intra-annual variability in the environment. Shifting towards generalist-dominated communities could have landscape-scale consequences for key soil processes. In this project, we impose differing environmental gradients in custom in-soil selection systems, allowing us to collect generalists and specialists of different types. We then assess the taxonomy and function of these assemblages. Ultimately, we aim to augment functional and ecological annotations of sequence data, and better inform microbial management.
Is there a role for microbial management in organic agriculture?
Funded by the USDA Organic Transitions Program (Lead PI is TH Bell; Co-PIs are Jason Kaye, Anna Busch)

(photo credit: Will King)
Many farmers believe soil microorganisms are key to soil functioning, but are unsure how to effectively manage microbial populations. Attempts to directly manage microorganisms are common, through approaches like compost development, Korean natural farming, and microbial product applications. However, the complexity and invisibility of microbial communities make it hard to benchmark potential solutions, leaving farmers without a framework for microbial management. In this project, we examine how soil type and farming practices interact with passive and active microbial management, in collaboration with farmer partners, and within a long-term organic project at Penn State. Specifically, we assess natural microbial colonization of farmed soils, the potential for forced microbial colonization of these soils, and the value of a widely used metric for assessing microbial health. We also interact with farmers through annual meetings, farmer conferences, and Extension publications.
Interactions between soil microbiomes and soil surface consortia

(photo credit: Ryan Trexler)
Funded by the Joint Genome Institute (Lead PI is TH Bell; Co-PI is Mary Ann Bruns)
We are assessing changes in the metatranscriptome of a Cyanobacteria-based soil surface consortium in the presence and absence of underlying soil microbiomes. This project aims to understand the degree to which in-soil competition can alter the function of microbial products for agriculture.
Root-centric microbiome analysis

(photo credit: Michela Centinari)
Funded by the USDA Foundational Program (Lead PI is Michela Centinari; Co-PIs are TH Bell, David Eissenstat)
We are examining the influence of root order, age, depth, and spatial arrangement on microbial recruitment. Whereas most studies of root-associated microorganisms use homogenized root systems, we seek to understand the fine-scale factors that shape root-microbe interactions at different points in space. The primary work for this project takes place in an experimental vineyard at Rock Springs in Pennsylvania, using root visualization windows within 1 metre deep root observation boxes. This field setup is unique, and allows us to sample grapevine roots across a substantial depth profile.
Selecting for desirable microbiome functions
Funded by the Atkinson Center for a Sustainable Future
 This project focused on the potential of root-associated microbiome breeding to influence Brassica rapa biomass in salt-amended and untreated soils, using replicated selection lines. We have also sequenced root-associated microbiomes at each generation to look for links between microbiome composition and B. rapa growth, and to determine the predictability of targeted microbiome selection.
This project focused on the potential of root-associated microbiome breeding to influence Brassica rapa biomass in salt-amended and untreated soils, using replicated selection lines. We have also sequenced root-associated microbiomes at each generation to look for links between microbiome composition and B. rapa growth, and to determine the predictability of targeted microbiome selection.
Relationships between soil contaminants, willows, and soil microbiomes
Funded by Genome Canada and FQRNT
I worked as a postdoctoral fellow on a large interdisciplinary project that aimed to enhance phytoremediation efficacy in willows. We used a variety of genomic approaches to examine the relationships between willows, microorganisms, soil characteristics, and contaminant uptake. These studies led us to understand that willow-microbe relationships vary in response to a number of factors, including willow species and soil contaminant concentrations. A better understanding of how plant-microbe relationships are influenced by the environment will allow us to characterize variability in the effectiveness of phytotechnologies. Ideally, this will eventually lead to more predictable outcomes when “green” technologies such as phytoremediation are applied, allowing them to better compete in the market with approaches that have a much larger environmental impact (e.g. dig and dump).
Predictability of microbiome composition and function following contamination
Supported by an NSERC PGS award

Many soil microbes can degrade petroleum, since they are adapted to metabolizing complex carbon compounds, such as those found in soil organic matter. This creates the potential for competition within the contaminated soil microbiome, just as we would be likely to find in most “pristine” microbial habitats. The focus of my Ph.D. work was the bioremediation of Arctic soils that have been contaminated by petroleum hydrocarbons. Nitrogen-based fertilizers are frequently added to hydrocarbon-contaminated soils, since they can allow increased microbial growth when carbon is in abundance and nitrogen is limiting, but not all hydrocarbon degraders are created equal. So which microorganisms benefit from this added nitrogen? What other factors might affect community composition in a contaminated soil? And are we promoting the most efficient hydrocarbon-degrading communities?
Impacts of winter warming on soil microbial function

(photo credit: Hugh Henry)
Our lab used a long-term field experiment to look at the effects of climate change on an old-field ecosystem. Infrared heaters warmed plots either year-round, or during the winter only, in an attempt to isolate the effects of climate change on winter processes. My role in this project was to examine how these factors affected microbial biomass, extracellular enzyme activity, and fungal:bacterial ratios. See here for more information about this project.