Protein homeostasis in plant organelles
Unlike prokaryotes, eukaryotic cells contain membrane bounded organelles; each performs some unique functions. The biogenesis and function of organelles are coordinated and tightly regulated by developmental and environmental cues so as to achieve their essential functions. Because of the membrane barrier, organelles like plastid, mitochondria and endoplasmic reticulum (ER) have their own independent protein quality control (PQC) systems inside to ensure protein folding, function and turnover in a timely manner. Compromised PQC in plant organelles often causes malfunction and impaired biogenesis of the organelles. Zhao laboratory is interested in understanding the mechanisms underlying protein homeostasis in plant organelles, with a focus on the role of heat shock family HSP90 proteins within plant ER and chloroplast.
In higher plants, there are four subfamilies of HSP90 proteins, the cytosolic HSP90A, ER-localized HSP90B, plastid-localized HSP90C and mitochondrial localized TRAP1-like protein. While HSP90 is not essential or absent in some prokaryotes, HSP90 is required for symbiont organelles as well as ER under either physiological or stress conditions. However, the role and mechanism of action of plant ER-localized HSP90B and plastid-localized HSP90C subfamilies are still elusive.
Function of chloroplast HSP90C in regulating thylakoid protein transport
Our lab has analyzed the Arabidopsis plastid HSP90C expression in different tissues and developmental stages and revealed its highly regulated expression during plant development. Down-regulation of HSP90C expression caused significant impairment of green tissue development and chloroplast maturation. HSP90C in the chloroplast stroma is critical for thylakoid membrane biogenesis likely through chaperoning proteins destined to thylakoid. From a proteomics screening analysis, we identified a cohort of HSP90C interactors including the PSII subunit PsbO1, an oxygen evolving complex subunit. It would be interesting to study the molecular mechanism by which HSP90C regulates thylakoid protein transport via the SEC translocase, by using the PsbO1 protein as a model client.

Function of plant ER-localized HSP90
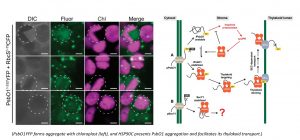
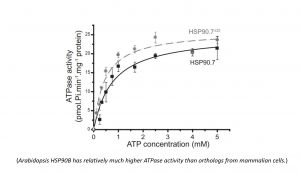
The HSP90 paralogues are also present in the endoplasmic reticulum of multiple-cell organisms. Different from cytosolic HSP90 proteins which normally works with a cohort of cochaperones and are involved in a large set of cytosolic protein homeostasis, the ER-located HSP90 isoforms are interacting with a much smaller set of proteins, though their functions are required for the development of multicellular organisms. Plant ER-localized HSP90B contains a unique charged sequence in the middle domain. This charged sequence could regulate the ATP-hydrolysis activity of HSP90B. Our group is particularly interested in unique plant ER-localized HSP90B interactors and cellular pathways which are dependent on HSP90B function, including but not limited to the those involved in protein secretion and the stem cell niche maintenance.
The mechanism of selective protein degradation by the proteasome
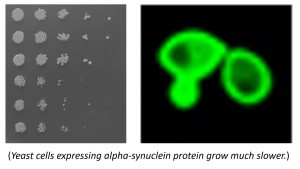
Neurodegenerative disorders such as Alzheimer’s disease, Parkinson’s disease and Huntington’s disease are caused by impaired neuronal function. A characteristic feature of neuronal cells, undergoing neurodegenerative disease, is accumulation of protein aggregates inside the cell. Aggregate accumulation is a result of defective protein turnover pathways. The ubiquitin-proteasome system within the cell selectively degrades proteins. The substrate selectivity is controlled primarily by the polyubiquitin pathway. However, under extreme and uncontrolled growth conditions, the proteasome can degrade proteins in unconventional ways depending on the stressors and the metabolic states of the cells. Our research group identified that the proteasome regulatory particle contains regions that can bind and trigger certain protein degradation independent of polyubiquitin pathway, particularly when the cellular metabolic status is changed (Paci et al, 2016). We hypothesized that the proteasome regulatory particle contains previously uncharacterized, but conserved features that can promote protein degradation without engaging the energy-inefficient polyubiquitination. Currently, native proteins from baking yeast cells and those that may account for the human neurodegenerative diseases, e.g. synuclein proteins, are being used as model substrates in the research project.
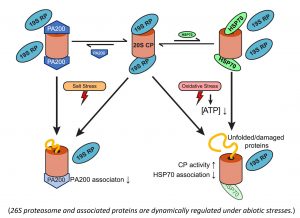
Other than the yeast model, we are also interested in the ubiquitin-independent protein degradation in other models like plant Arabidopsis. It is expected ubiquitin-independent protein degradation by either the 20S or 26S proteasome may play critical roles in certain developmental stage of plants, and it would be interesting to understand how the 26S proteasome holoenzyme activity is regulated in plant under stress conditions. For example, we have observed that molecular chaperone HSP70 is normally associated with plant 20S proteasome but gets less associated under the oxidative stress condition.