
Welcome to the Weir Lab at the University of Toronto. Our lab uses genomic and comparative methods to investigate the pace of evolution in the tropics versus high latitudes. The tropics have exceptional levels of biodiversity, and understanding the drivers of that diversity is an underlying theme in our lab’s research. As lab members we are also committed to sustainable lifestyle choices that are necessary for the biodiversity we study to thrive on our planet (see Biodiversity and Sustainable Living). Using birds as our model group, we employ a variety of genomic, field and computational approaches to estimate evolutionary rates across latitudinal gradients.
The Weir Lab is proud to collaborate with excellent colleagues in New Zealand, Brazil, and Israel (Hebrew University).
LAB AWARDS
- Matthew Thorstensen receives the 2025 Eric and Wendy Schmidt AI in Science Postdoctoral Fellowship
- Gihyun Yoo receives a 2025 NSERC CGSD Fellowship
- Else Mikkelsen receives the 2024 CSEE Excellence in Doctoral Research award
- Else Mikkelsen receives the 2024 Abrams Prize awarded by EEB
- Ellen Nikelski receives a 2022 NSERC CGSD Fellowship
- Sean Anderson receives our department's best graduate student paper of 2023 award (read.here)
- Jordan Bemmels receives our department's best postdoc paper of 2022 award (read.here)
- Sean Anderson receives the 2021 Abrams Prize awarded by EEB
- Jason Weir receives the 2021 International Biogeographic Society's MacArthur & Wilson Award
The Genomics of Speciation
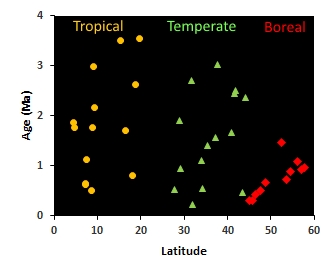
Our lab investigates how reproductive isolation accumulates through time, with a focus on comparing the tempo and genetic architecture of speciation in tropical versus boreal birds. A central goal is to understand the speciation continuum—the gradual progression from initial divergence to complete isolation—by examining avian hybrid zones that span a wide range of divergence times and ecological settings.
Our published work has shown that tropical bird species can maintain hybrid zones for up to ~4 million years post-divergence (see figure to the right), while in boreal regions such zones typically persist for less than ~1.5 million years (Weir & Price 2011; Weir et al. 2015), suggesting accelerated rates at which premating isolation is completed at high latitudes. We have also demonstrated that Amazonian rivers are often semi-permeable barriers (Weir et al. 2024), with many species pairs forming hybrid zones in adjacent headwater regions (Weir et al. 2015; Pulido-Santacruz et al. 2018; Cronemberger et al. 2020; Barrera-Guzmán et al. 2022; Weir et al. 2024). These patterns suggest that speciation proceeds more slowly in the tropics, potentially due to extended periods of gene flow and more gradual accumulation of reproductive barriers.
We are leveraging a dataset of over 40 de novo avian reference genomes for closely related pairs of species of different ages to examine how mitonuclear incompatibilities accumulate over evolutionary time. By comparing patterns of mitochondrial–nuclear coevolution across tropical and temperate lineages, we aim to uncover when mismatches arise between mitochondrial and nuclear-encoded genes and how these contribute to the buildup of postzygotic isolation. Together, these complementary approaches allow us to reconstruct both the genomic and temporal dynamics of speciation across contrasting environments.
Introgression and hybrid speciation

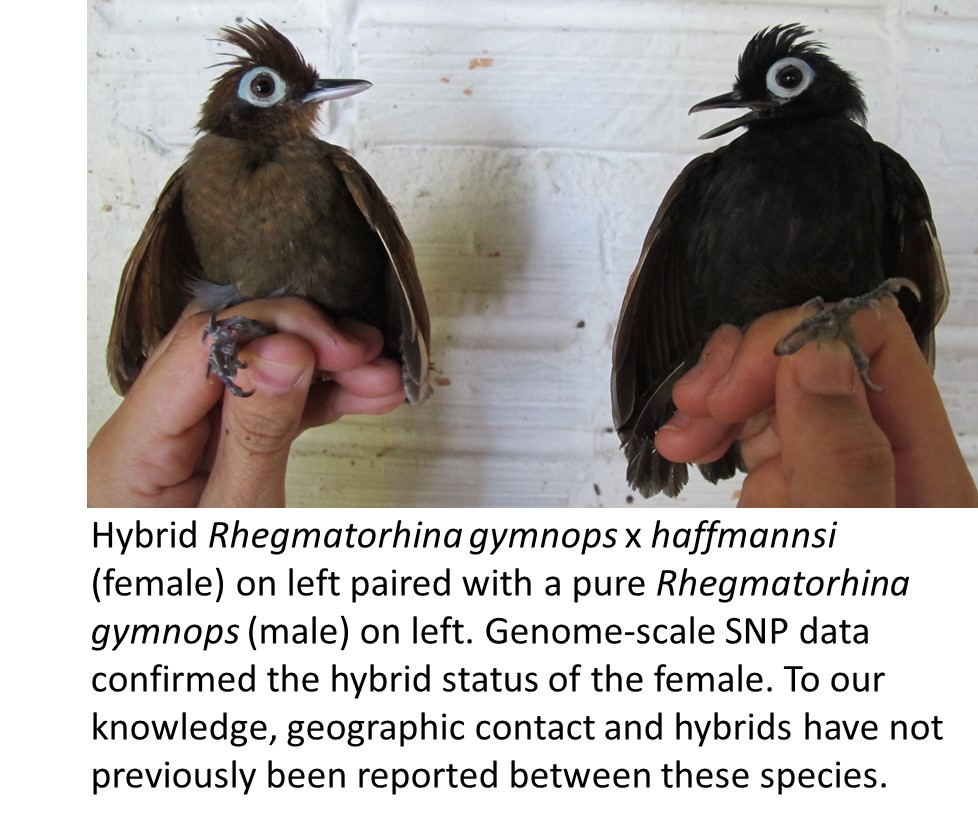
Hybridization can also generate new species. We recently showed that the Golden-crowned Manakin likely originated ~180,000 years ago from hybridization between Snow-capped and Opal-crowned Manakins (Pulido-Santacruz et al. 2018). This is the first known avian hybrid species from the Amazon, and its phenotype is not intermediate between its parent species. Else Mikkelsen is now using whole-genome sequencing to confirm this hybrid origin. Her work has also revealed mitogenomic capture and nuclear introgression from Pomarine Jaeger into Great Skua (Systematic Biology, 2023), and she is uncovering chromosomal inversions that appear to have spread across multiple species boundaries via introgression.
Phylogenetic estimates of speciation and extinction rates
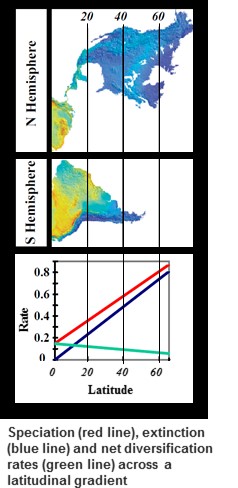
Are speciation rates fastest in the tropics, and can they explain the high species richness there? Using phylogenetic data we showed that rates of speciation have been much faster at high latitudes, and slower in the tropics over the past few million years. In contrast, extinction rates were very slow in the tropics, and might explain how the tropics have been able to accumulate such high levels of species richness (read here). Recent research by former Ph.D. student Dr. Paola Pulido-Santacruz extended this work across the entire bird phylogeny. Her work suggests that extinction rates are consistently lower in geographic regions with low species richness across many replicate orders of birds, while speciation rates showed no consistent pattern with latitude across replicate clades (read here). Together, these studies suggest that low tropical extinction could be the key historical driver behind the latituidnal diversity gradient.
Latitudinal gradients in trait evolution
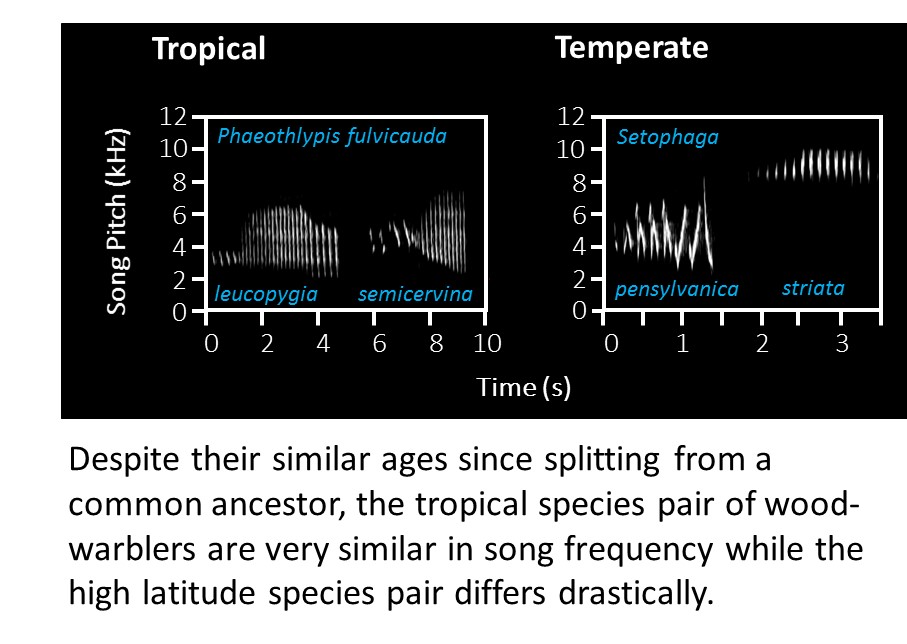
We investigate whether rates of evolution vary across latitudinal and ecological gradients in traits (song, colour, morphometrics and climatic niche) important for speciation and ecological differentiation. A key focus has been on rates of evolution in the courtship songs used by birds to attract mates. Our results demonstrate faster evolution of bird song (Weir & Wheatcroft 2011, Proc. Roy B; Weir et al. 2012 Evolution. Former MSc student Adam Lawson demonstrated a much faster rate of climatic niche evolution at high latitudes, suggesting that divergent selection pressures are stronger towards the poles (Lawson & Weir 2014, Ecology Letters) while PhD student Vanessa Luzuriaga-Aveiga has demonstrated that increased elevational differentiation results in increased rates of song and morphometric trait evolution across the Amazon and eastern Andean slopes (Luzuriaga-Aveiga & Weir 2019, Ecology Letters). PhD student Sean Anderson has developed new models of trait evolution (and a new R package) that quantify the degree of divergent adaptation (Anderson & Weir 2020, Am. Nat.) and more recently that distinguish between character displacement,
Comparative Phylogeography and Biogeography

An ongoing focus of the lab is to utilize comparative approaches across many species to address key biogeographic questions such as the role of ice sheets in driving allopatric speciation in boreal birds (read.here) and New Zealand kiwi (read here), the role of climate change in driving diversification of lowland and highland birds of the Neotropics (read here), and the role of landbridge completion in driving the Great American Biotic Interchange between North and South American birds (read here). We are using genome-wide and whole genome datasets to study phylogeographic questions in a variety of other North American and Amazonian species: manakins (Ph.D. student Else Mikkelsen and former student Dr. Alfredo Barrera-Guzman), antbirds (former students Dr. Aurea Cronemberger and Dr. Maya Faccio), tanagers (Ph.D. student Vanessa Luzuriaga), flyctachers (former post doc Dr. Jordan Bemmels), warblers, thrushes (with collaborators Alyssa FitzGerald and Jeremy Kirchmam), titmice (with collaborator Claire Curry), and kiwi (with former post doc Dr. Jordan Bemmels).
Weir Lab Opportunities (details here)
